Accession
MI0009819
Description
Bos taurus
bta-mir-378-1 precursor miRNA
Literature search
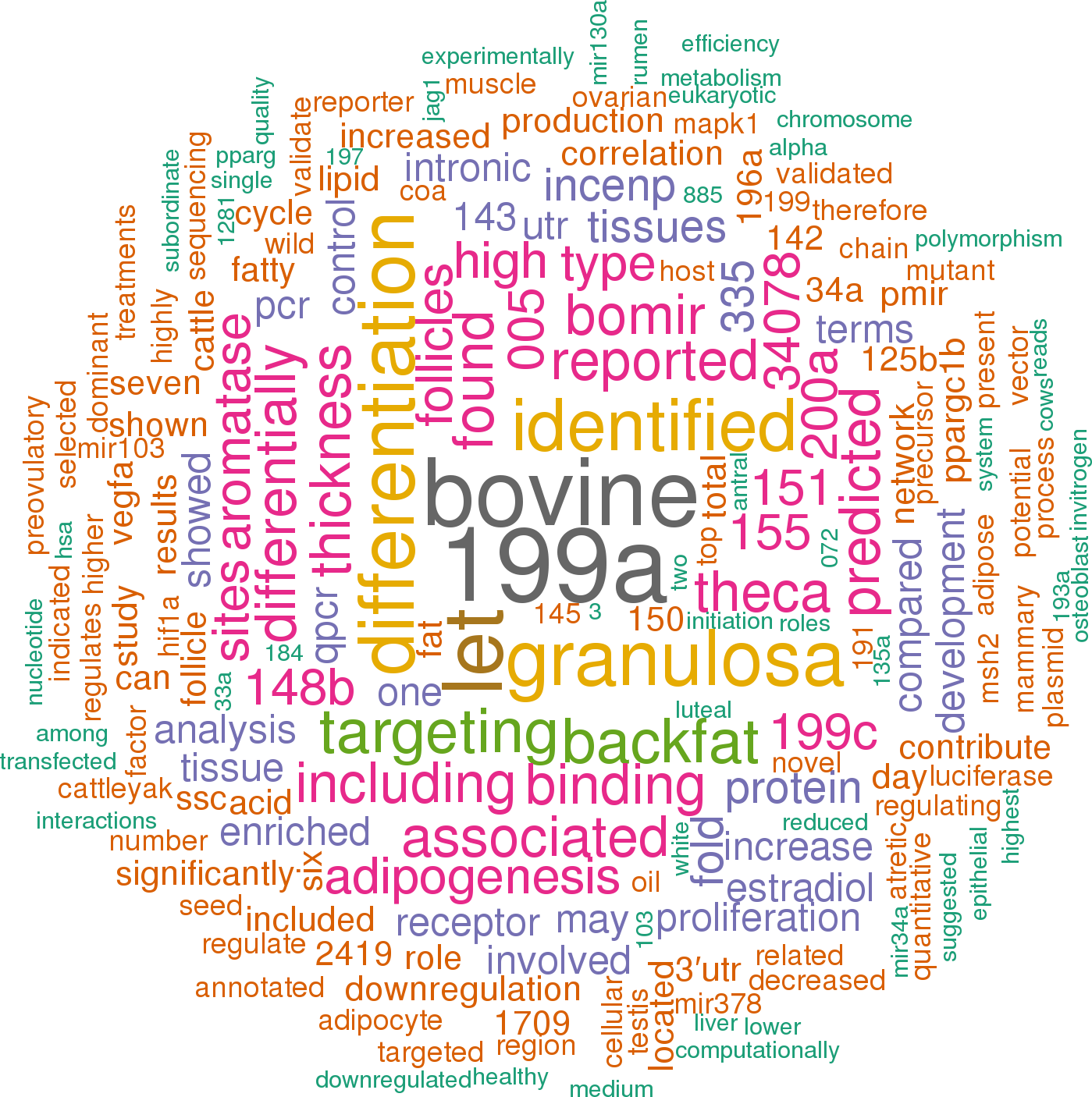
24 open access papers mention bta-mir-378-1
(101 sentences)
(101 sentences)
Sequence
535504
reads,
4317
reads per million, 77 experiments
agggcuccugacuccagguccuguguguuaccucgaaauagcACUGGACUUGGAGUCAGAAGGCcu
(((.((.((((((((((((((.((((..(........)..)))).)))))))))))))).)).)))
(((.((.((((((((((((((.((((..(........)..)))).)))))))))))))).)).)))
Structure
g c u gu acc agg cu cugacuccaggucc gugu u u ||| || |||||||||||||| |||| | ucC GA GACUGAGGUUCAGG CAcg a c G A U au aag
Annotation confidence
High
Do you think this miRNA is real?
Genome context
chr7: 63343197-63343262 [+]
Mature bta-miR-378
| Accession | MIMAT0009305 |
| Description | Bos taurus bta-miR-378 mature miRNA |
| Sequence | 43 - ACUGGACUUGGAGUCAGAAGGC - 64 |
| Evidence |
experimental
cloned [1] |
References
|



