Accession
MI0000988
Description
Arabidopsis thaliana
ath-MIR169n precursor miRNA
Gene family
MIPF0000012;
MIR169_1
Literature search
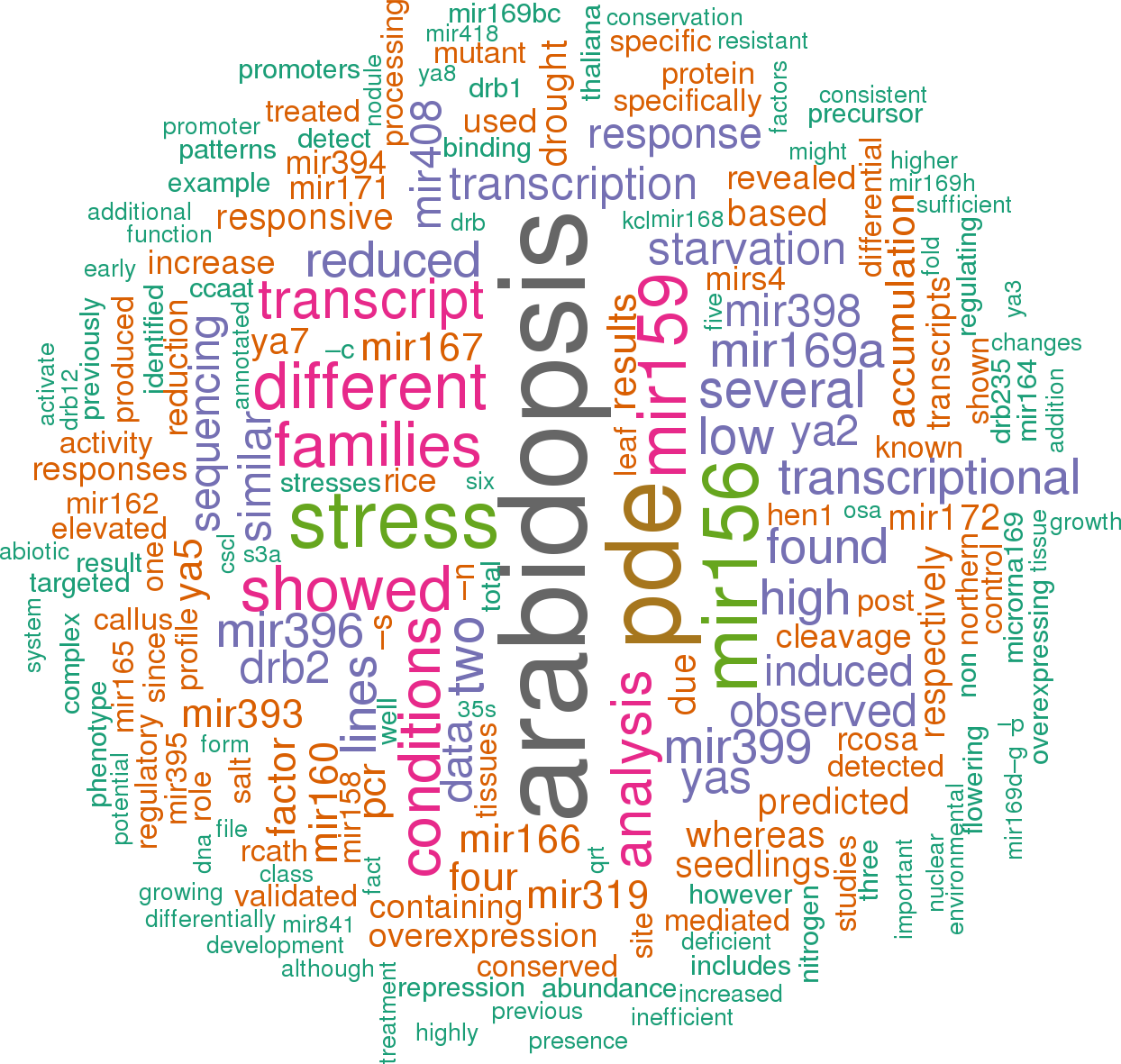
34 open access papers mention ath-MIR169n
(159 sentences)
(159 sentences)
Sequence
gaugaagaagagaggucuaacauggcggaaagcgucauguuuagUAGCCAAGGAUGACUUGCCUGaucuuuuucgccuccacgauucaauuucaaauucaugcauuuuggauuauuauaccuuuuaaaguauaauaggucaaauaucauguugaaucuugcggguuagguuucaggcagucucuuuggcuaucuugacaugcuuuuuccauccau
((((((((((....(((.(((((((((.....)))))))))..(((((((((((.((((.((((((....(((.((((.((.(((((((.........((((...(((((.(((((((((........))))))))).)))))...))))))))))).)).)))).)))..)))))))))))))))))))))...)))...)))))).))))...
((((((((((....(((.(((((((((.....)))))))))..(((((((((((.((((.((((((....(((.((((.((.(((((((.........((((...(((((.(((((((((........))))))))).)))))...))))))))))).)).)))).)))..)))))))))))))))))))))...)))...)))))).))))...
Structure
--- - agag uaacauggcggaaagcgucauguuua U U ucuu c c c uuucaaauu cau a cuu gaug aagaag guc gUAGCCAAGGA GACU GCCUGa uuu gccu ca gauucaa caug uuugg uuauuauac u |||| |||||| ||| ||||||||||| |||| |||||| ||| |||| || ||||||| |||| ||||| ||||||||| cuac uuuuuc cag uaucgguuucu cuga cggacu gga uggg gu cuaaguu guac aaacu gauaauaug u uac c -gua -----------------------uuc - - --uu u c u --------- uau g aaa
Annotation confidence
Not enough data
Do you think this miRNA is real?
Comments
This sequence is a predicted paralogue of the previously identified miR169 family [1], later experimentally verified [2]. It is predicted to target mRNAs coding for the CCAAT Binding Factor (CBF) and HAP2-like transcription factors.
Genome context
chr3: 9878540-9878754 [-]
Clustered miRNAs
5 other miRNAs are < 10 kb from ath-MIR169n
| Name | Accession | Chromosome | Start | End | Strand | Confidence |
|---|
Mature ath-miR169n
| Accession | MIMAT0000919 |
| Description | Arabidopsis thaliana ath-miR169n mature miRNA |
| Sequence | 45 - UAGCCAAGGAUGACUUGCCUG - 65 |
| Evidence |
experimental
5'RACE [2], cloned [2], 454 [3-4], MPSS [3], Illumina [5] |
References
|

